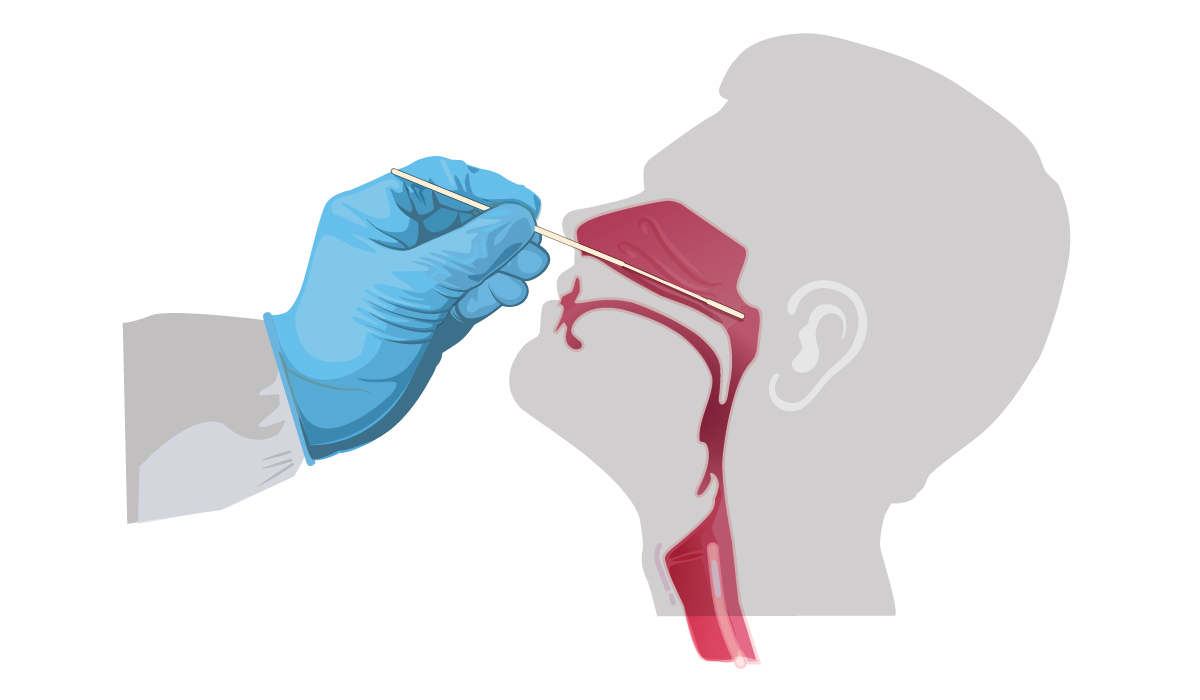
08 Apr THE TEST DETECTING SARS-CoV-2
Most tests for detecting SARS-CoV-2 are based on one of the most -if not the most- used technique in molecular biology; namely PCR. PCR (Polymerase Chain Reaction) can amplify (make many copies of) a short region (e.g. a gene) of a DNA molecule. Why do we need to amplify a gene? Simply because in its original state, it is not enough to be detected and further analyzed.
Scientists know the exact genetic code (i.e. the sequence) of SARS-CoV-2 and this allows them to design the appropriate ‘primers’ (very short segments complementary to the desired SARS-CoV-2 gene) which can bind to the gene once they encounter it. For the purposes of viral DNA detection, qPCR is used, which uses a fluorescent label in order to allow scientists to detect and quantify the end product. The concept is based on the usage of a fluorescent dye that does not emit significant amount of fluorescence in the presence of single-stranded DNA. As the reaction of DNA amplification progresses, more and more double-stranded products – specific of a viral gene – are generated. The fluorescent dye intercalates into these products and begins to fluoresce.
The fluorescence signal is directly proportional to DNA concentration over a broad range, and the linear correlation between PCR product and fluorescence intensity is used to calculate the amount of DNA present at the beginning of the reaction. In other words, analyzing the fluorescent intensity determines whether the gene of SARS-CoV-2 is truly (positive result) in the sample drawn by a patient or not (negative result).
In the case of SARS-CoV-2, the genetic material of the virus is RNA. As a result, it is necessary to ‘transcribe’ (change) the RNA into DNA via reverse-transcriptase, before PCR can be run (since PCR works by making copies of DNA). The question that might arise here is how do we extract RNA from an organism, tissue, or a cell. The principle can be described here: link
DIAGRAMMATICALLY: FROM THE PATIENT TO THE TEST
1. Patient provides sample for analysis. Photo source: https://www.cbc.ca
2. RNA is extracted by the sample![]()
3. RNA is transcribed (transformed) into DNA in order to be amplified by PCR![]()
4. After enough copies of viral DNA are made, it can be clear whether the patient is positive or negative to the viral DNA.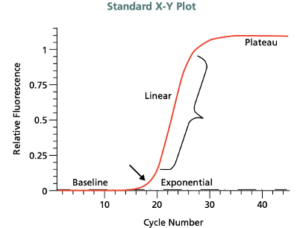
PCR Amplification Plot: Adapted from: https://www.gene-quantification.de/SIAL-qPCR-Technical-Guide.pdf A PCR consists of a series of 20–40 repeated temperature changes, called cycles and shown on the x axis. The DNA should ideally double in every cycle. The higher the amount of the initial sample DNA, the sooner (early PCR cycles) the accumulated product is detected in the fluorescence plot. The doubling of genetic material per cycle makes PCR a geometric amplification. Therefore, the linear plot of the data should show a classic exponential amplification in the standard X-Y plot.



Sorry, the comment form is closed at this time.